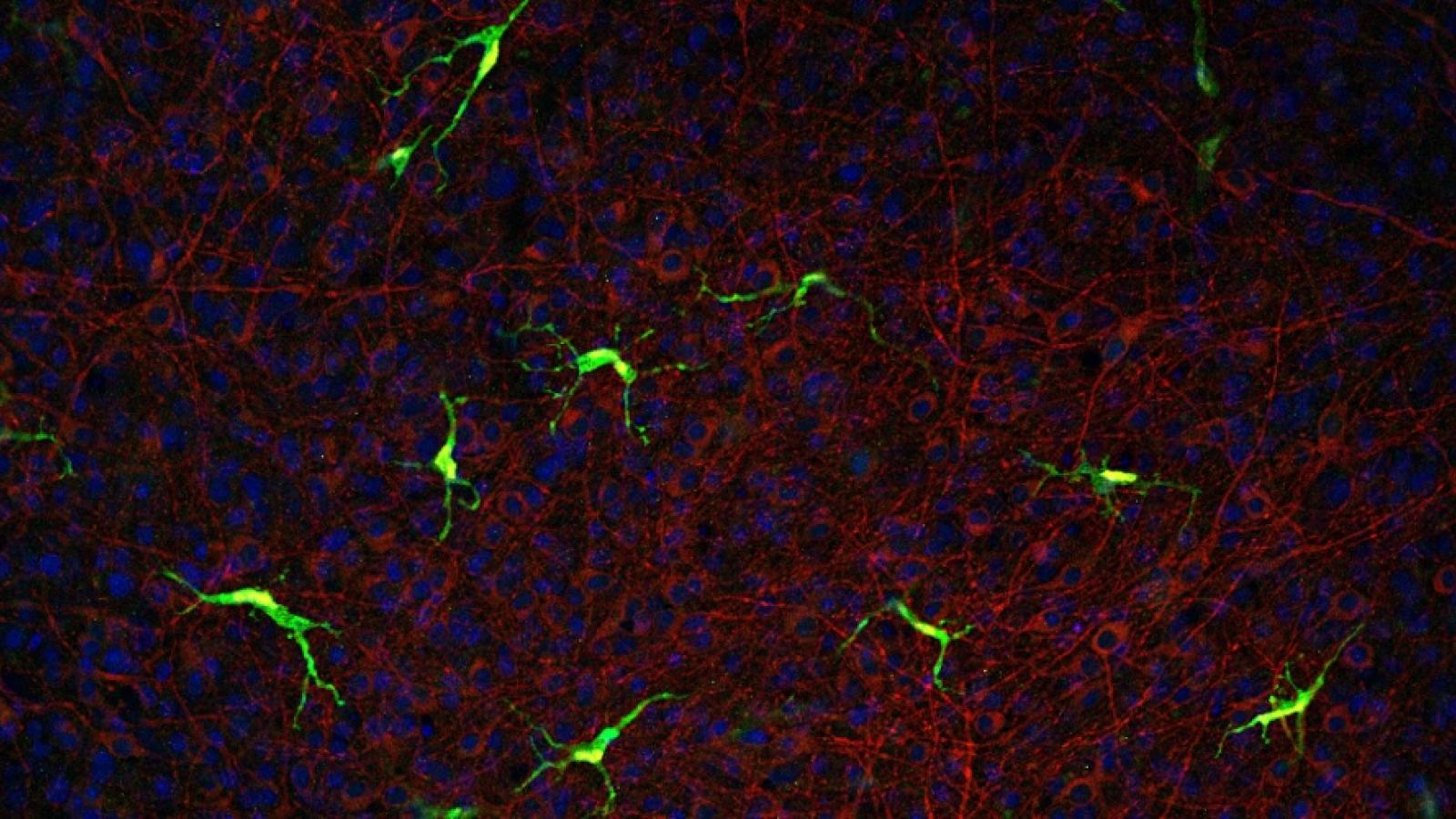
The iPSC Platform to Model Alzheimer’s Disease Risk (IPMAR), led by the UK DRI at Cardiff, is a major resource to study Alzheimer’s risk, aiming to help researchers worldwide explore factors that contribute to development of the condition. Led by Prof Julie Williams, the IPMAR team have created a panel of cell lines from people with varying risk for late- and early-onset Alzheimer's. A paper detailing the resource and cell lines available has been published in Stem Cell Reports. We spoke to Dr Hazel Hall-Roberts, IPMAR Stem Cell Lead, to find out more.
Hi Hazel! Could you introduce us to IPMAR and its aims?
IPMAR is a collection of over 100 iPSC lines that capture the effects of genes that contribute to the development of sporadic Alzheimer’s. Our aim was to make a well-characterised iPSC platform to better represent the common genetic forms of Alzheimer’s, available to all researchers. We wanted to enable study of the fundamental molecular and cellular processes underlying sporadic Alzheimer’s, and provide a platform for drug discovery. The iPSCs were made using our existing Alzheimer’s cohort in Cardiff. In the Cardiff AD cohort, collected over the last 30 years, we have over 4000 Alzheimer’s disease participants and over 2000 control participants, from whom we have collected a wealth of data including cognitive measures, genetic profiling and biological samples.
Late-onset Alzheimer’s disease is a multifactorial and extremely complex condition. In the majority of cases, the risk of developing it is determined by thousands of small contributions from our genes, along with environmental factors such as lack of exercise. Researchers have been analysing huge genetic datasets to generate polygenic risk scores (PRS), which are a calculation of an individual’s overall risk for developing Alzheimer’s.
We selected study participants with Alzheimer’s (both late- and early-onset) that had extremely high polygenic risk for the disease, including in some specific pathways such as complement, an area of the innate immune system (led by Dr Sarah Carpanini). Our controls are healthy aged participants with extremely low polygenic risk, so that we are comparing two extremes to maximise the effect size of any potential phenotypes.
Did IPMAR do what it set out to do?
We started in January 2021, so the pandemic caused us some initial delays as we struggled to obtain the materials we needed in that first year. Resuscitating 20-year old frozen cells and creating iPSCs from them was a significant technical challenge to overcome, and we found that we had to thaw a lot more samples than we used! However, we achieved our aim of generating around 100 iPSC lines. In collaboration with Dr Sally Cowley at the University of Oxford, we created 53 iPSC lines, and then a further 56 iPSC lines were generated by Oxford Stem Tech using similar methodology.
What is shown in the new Stem Cell Reports paper?
The paper is a ‘resource article’, so we present a detailed description of the iPSC resource and the methods that we used for participant selection, iPSC generation and quality control. We hope that other researchers will look at what we have and plan future projects using the lines.
What has been the impact of the project?
Many of the IPMAR lines have already been distributed to other researchers as part of collaborations to study the phenotypes of differentiated brain cell types, and we’re expecting the first results to be published this year, including our own microglia study. We anticipate that the results of these studies, which include ‘omics data, will inform researchers of the early cellular and molecular dysfunction commonly experienced by people with common genetic forms of Alzheimer’s.
How can others access the data/databases?
For iPSC access, we welcome collaborations and can directly provide lines under an agreement.
- For the global polygenic risk lines, direct enquiries to me (hazel.hall-roberts@ukdri.ac.uk).
- For the complement pathway-specific lines, contact Sarah Carpanini (sarah.carpanini@ukdri.ac.uk).
- 20 iPSC lines are currently available from the EBiSC biobank.
- We’re making the QC data for the iPSC available at https://hPSCreg.eu.
- There is a rich dataset associated with the iPSC donors that includes clinical, cognitive and non-cognitive longitudinal data and genome-wide microarray data, which people can apply to access from the Dementias Platform UK Data Portal.
What’s next for the project?
Later this year we will publish a detailed characterisation of iPSC-microglia differentiated from the late-onset Alzheimer’s IPMAR lines. We hope that the future will bring more collaborations with other research groups, who will have fresh ideas and expertise in other brain cell types, culture systems or analysis techniques.
Reference: Maguire E, Winston J, Ellwood SH, O'Donoghue R, Shaw B, Morales AC, Keat S, Evans A, Marshall R, Luckcuck L, Brown L, Salis E, Leonenko G, Denning N; EADB consortium; Allen ND, Escott-Price V, Webber C, Taylor PR, Sims R, Cowley SA, Williams J, Carpanini SM, Hall-Roberts H. Modeling common Alzheimer's disease with high and low polygenic risk in human iPSC: A large-scale research resource. Stem Cell Reports. 2025 Jun 21:102570. doi: 10.1016/j.stemcr.2025.102570.